Spatial Transcriptomics Technologies
Evolution of Spatial Transcriptomics (2016-2025)
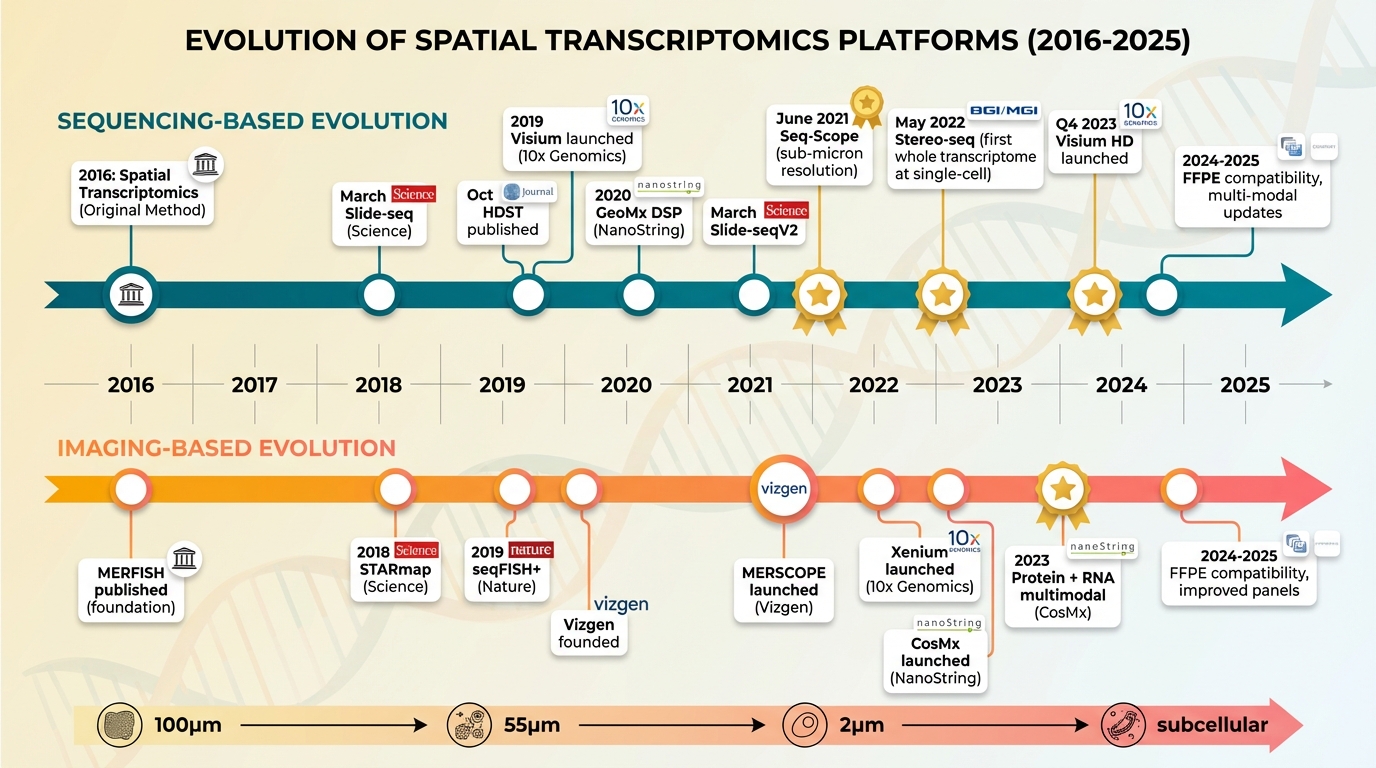
Figure 1. Timeline of spatial transcriptomics technology development. Upper track (teal): sequencing-based methods from original ST (2016) through Visium, Slide-seq, Seq-Scope (2021), Stereo-seq (2022), to Visium HD (2023). Lower track (orange-coral): imaging-based methods from MERFISH foundation through STARmap, seqFISH+, MERSCOPE, Xenium, and CosMx. Gold stars indicate breakthroughs. Bottom scale shows resolution progression from 100μm to subcellular.
What is Spatial Transcriptomics?
Spatial transcriptomics refers to a collection of methods that measure gene expression while preserving the spatial location of transcripts within tissues. Unlike traditional RNA-seq that requires tissue homogenization (losing spatial information) or single-cell RNA-seq that requires cell dissociation (losing tissue context), spatial transcriptomics maintains the native tissue architecture while profiling the transcriptome.
Why Spatial Context Matters
- Tissue Architecture Understanding: Cells behave differently based on their location - a cancer cell at the tumor edge behaves differently than one in the center
- Cell-Cell Interactions: Understanding signaling between neighboring cells requires knowing their spatial relationships (ligand-receptor pairs)
- Microenvironment Mapping: Immune cells, stromal cells, and parenchymal cells form functional niches that can only be studied in spatial context
- Disease Heterogeneity: Tumors, inflammatory lesions, and developmental abnormalities show spatial patterns that inform diagnosis and treatment
- 3D Organization: Tissues are three-dimensional; understanding cell clustering and connectivity patterns requires volumetric analysis
Three Major Technology Categories
1. Sequencing-Based Methods (Capture + Seq)
Use spatially-barcoded oligonucleotides to capture mRNA at defined positions, followed by next-generation sequencing. Offer whole-transcriptome coverage but lower spatial resolution.
2. Imaging-Based Methods (In Situ)
Detect transcripts directly in tissue using fluorescence hybridization or in situ sequencing. Achieve single-molecule/subcellular resolution but typically target specific gene panels.
3. Emerging & Multi-Modal Methods
Novel approaches combining transcriptomics with other modalities (protein, translation, temporal dynamics) or using innovative readout strategies.
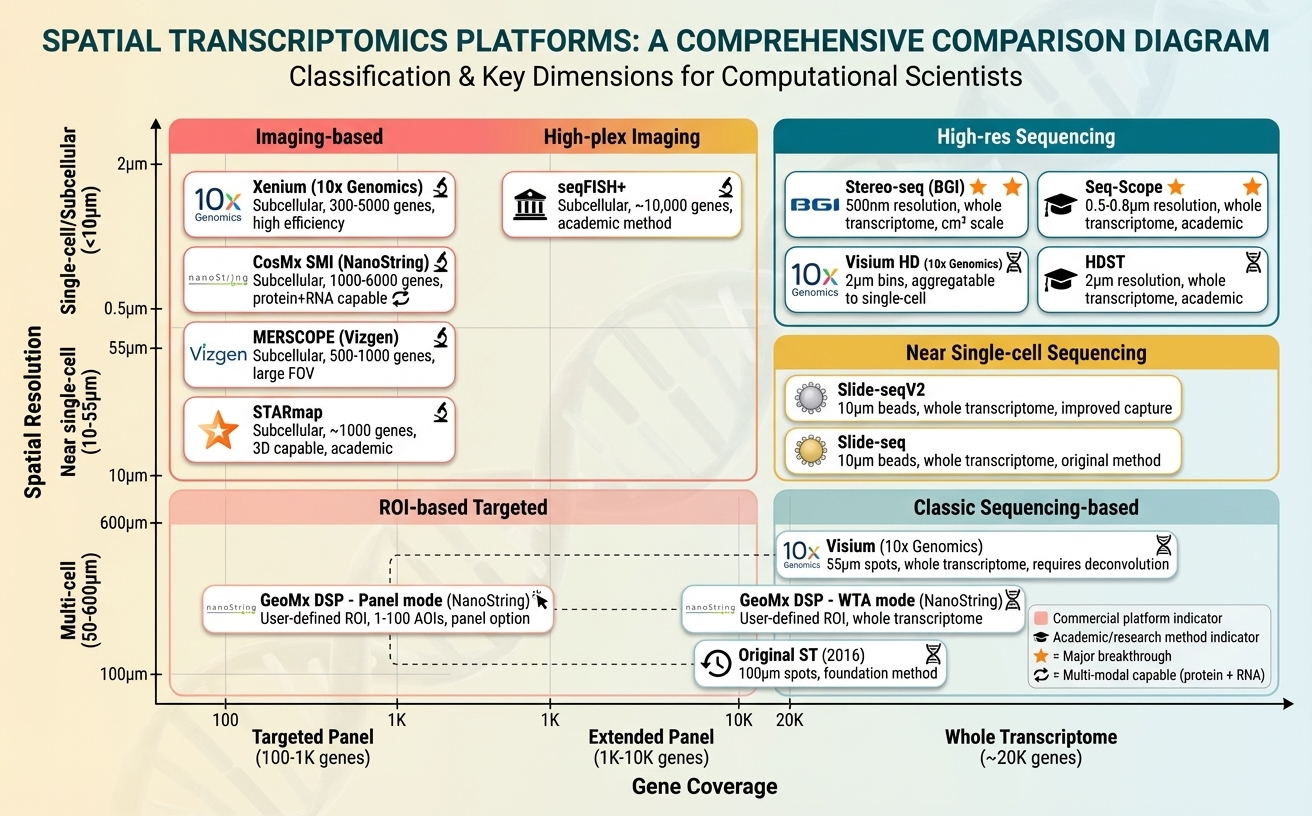
Figure 2. Platform classification by spatial resolution (y-axis) and gene coverage (x-axis). Upper left (coral): imaging-based platforms (Xenium, CosMx, MERSCOPE, STARmap) with subcellular resolution and targeted panels. Upper right (teal): high-resolution sequencing (Stereo-seq, Seq-Scope, Visium HD, HDST) with whole transcriptome. Middle right (golden): near single-cell sequencing (Slide-seqV2). Lower right (light teal): classic sequencing (Visium, GeoMx WTA). Lower left: ROI-based targeted (GeoMx panel mode). Stars denote subcellular + whole transcriptome breakthroughs.
Technology Comparison Guide
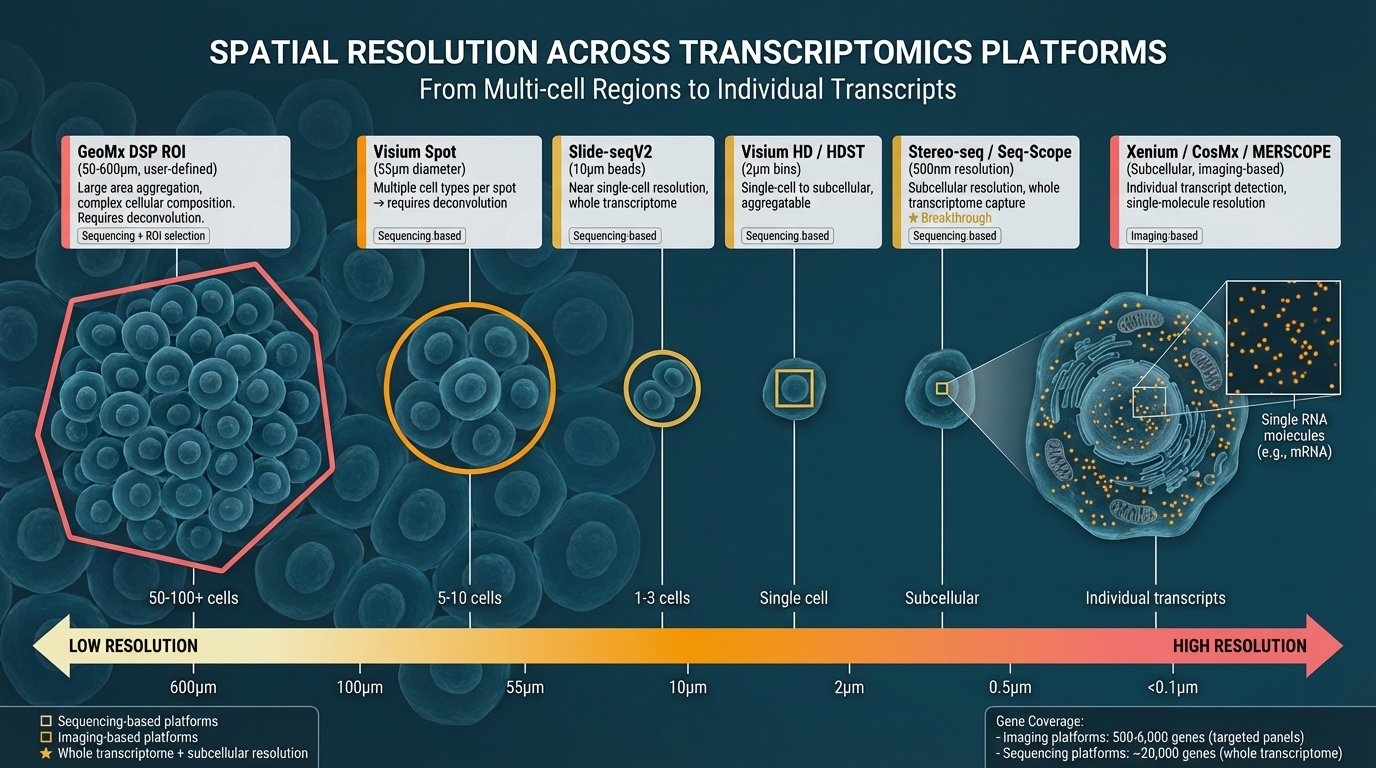
Figure 3. Visual comparison of spatial resolution from multi-cell regions to individual transcripts. Left to right: GeoMx DSP ROI (50-600μm, 50-100+ cells), Visium (55μm, 5-10 cells), Slide-seqV2 (10μm, 1-3 cells), Visium HD/HDST (2μm, single-cell), Stereo-seq/Seq-Scope (0.5μm, subcellular, ★), Xenium/CosMx/MERSCOPE (<0.1μm, individual transcripts). Yellow outlines: sequencing-based; coral outlines: imaging-based. Sequencing platforms capture ~20K genes; imaging platforms detect 500-6,000 genes.
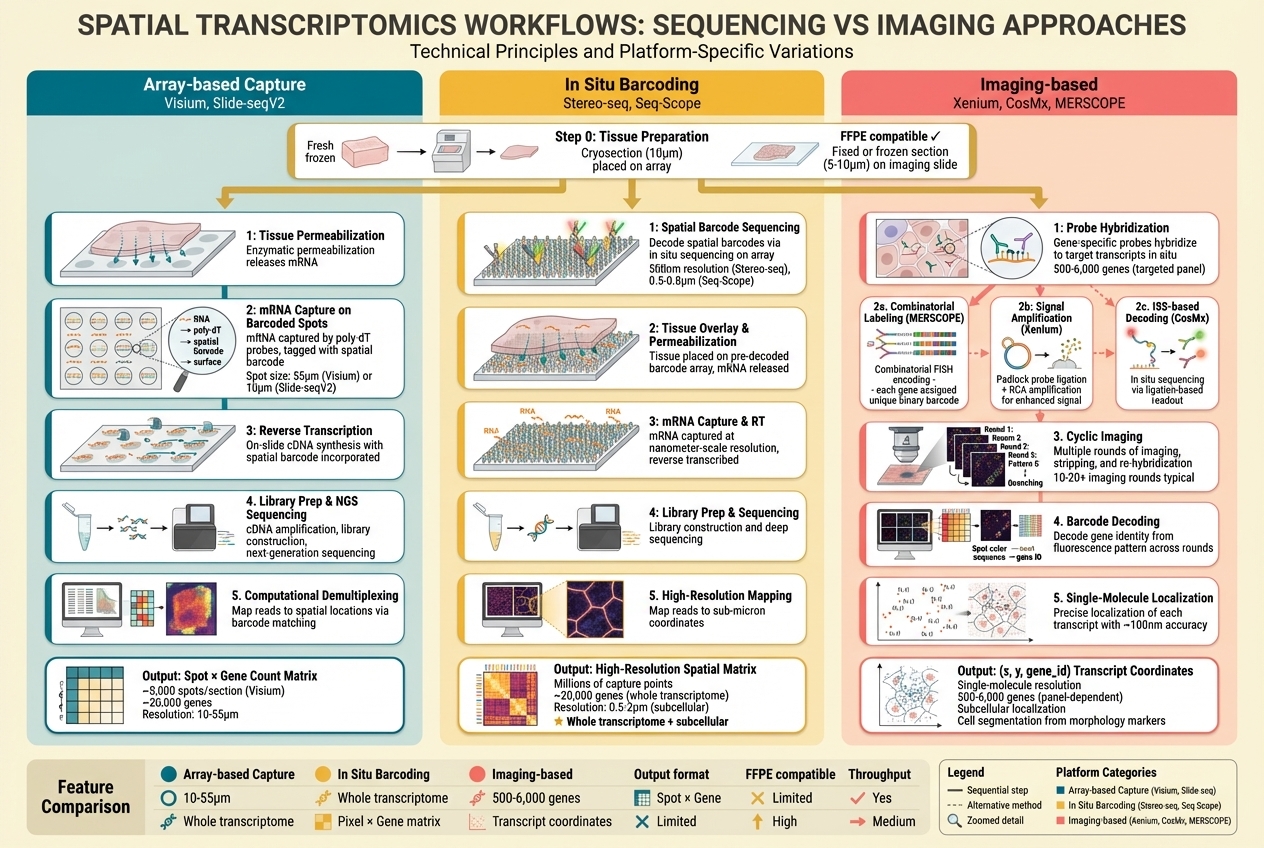
Figure 4. Technical workflows across three approaches. Left (Array-based Capture): tissue permeabilization → mRNA capture on barcoded spots → reverse transcription → library prep → demultiplexing; output: Spot × Gene matrix. Center (In Situ Barcoding): spatial barcode sequencing → tissue overlay → mRNA capture → sequencing → high-resolution mapping; output: subcellular spatial matrix (★). Right (Imaging-based): probe hybridization → platform-specific detection (combinatorial/RCA/ISS) → cyclic imaging → barcode decoding → single-molecule localization; output: (x,y,gene_id) coordinates. Bottom table compares resolution, genes, output format, FFPE compatibility, and throughput.
Resolution vs Coverage Trade-off
| Method | Resolution | Gene Coverage | Tissue Depth | Best For |
|---|---|---|---|---|
| Visium | 55 um spots | Whole transcriptome | 10 um sections | Discovery, FFPE tissues |
| VisiumHD | 2-8 um bins | Whole transcriptome | 10 um sections | High-resolution discovery |
| Slide-seq V2 | 10 um beads | Whole transcriptome | 10 um sections | Near-cellular resolution |
| Stereo-seq | ~500 nm | Whole transcriptome | Thin sections | Subcellular patterns |
| Xenium | Subcellular | 100-5000 genes | 5-10 um | Targeted, clinical FFPE |
| seqFISH+ | Super-resolution | 10,000 genes | ~5 um | Discovery + localization |
| CosMx SMI | Subcellular | 1000+ genes | 5 um | High-plex imaging |
| Deep-STARmap | Single-cell | 1000+ genes | 60-200 um | 3D architecture |
| GeoMx DSP | ROI-based | Whole transcriptome | 10 um | Targeted enrichment |
Choosing the Right Technology
For Discovery Studies (Unbiased)
Use sequencing-based methods (Visium, VisiumHD, Slide-seq, Stereo-seq) when you need whole-transcriptome coverage without predefined gene targets. Best for hypothesis generation and atlas building.
For Targeted Validation (High Resolution)
Use imaging-based methods (Xenium, CosMx, seqFISH+, MERFISH) when you have specific genes of interest and need single-cell or subcellular resolution. Best for validating scRNA-seq findings.
For 3D Tissue Architecture
Use Deep-STARmap/RIBOmap for thick tissue (60-200 um) profiling to capture complete 3D cellular organization. Essential for studying cell-cell interactions and spatial clustering that are missed in 2D.
For Clinical FFPE Samples
Use Visium, Xenium, or GeoMx — platforms optimized for archival formalin-fixed paraffin-embedded tissue.
Quality Check: Assess RNA integrity using DV200 (% of RNA fragments >200 nucleotides). Aim for DV200 ≥50% for optimal results.
For Multi-Modal Integration
Use SPOTS for simultaneous protein + transcriptome, Deep-RIBOmap for transcription + translation, or combine Xenium + Visium + Chromium for comprehensive multi-platform profiling.
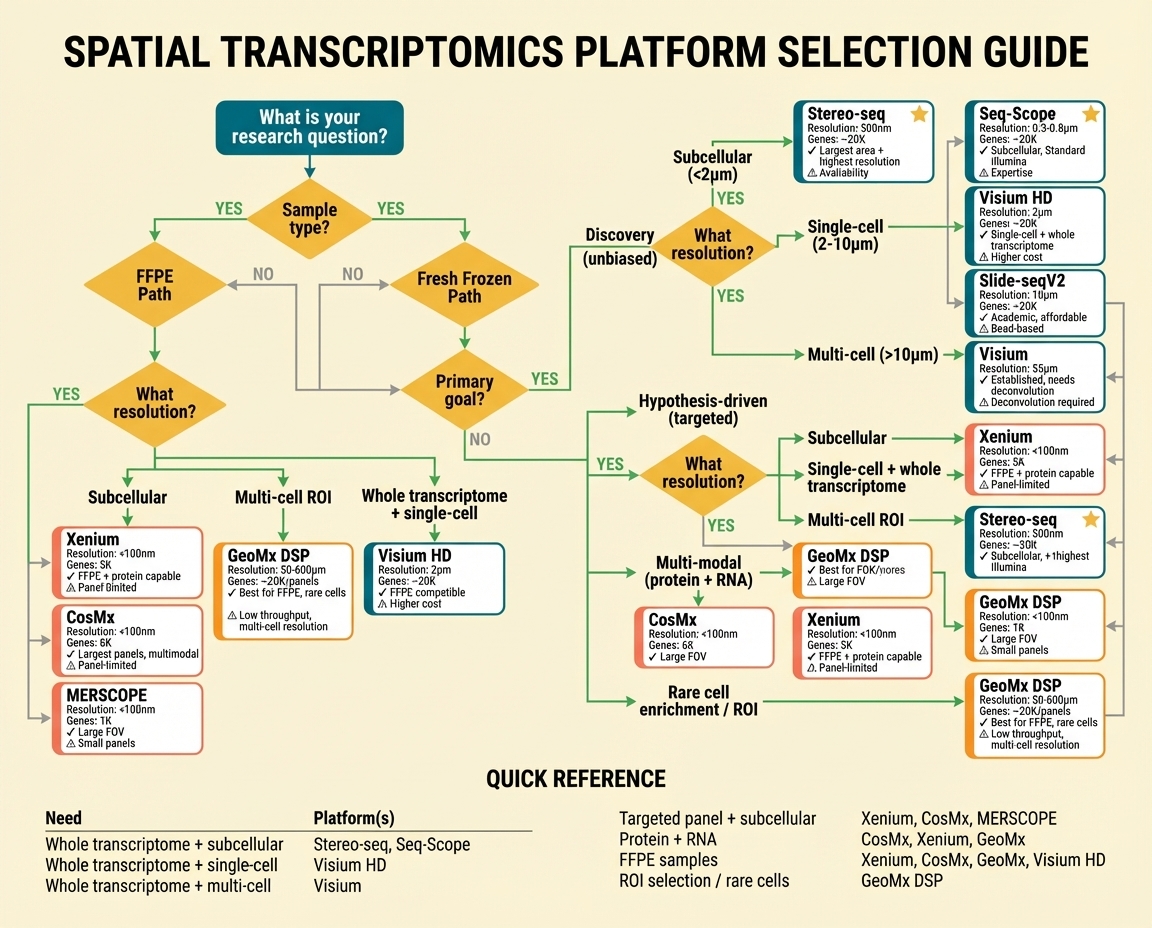
Figure 5. Decision flowchart for platform selection. Starting from sample type: FFPE samples → subcellular (Xenium, CosMx, MERSCOPE), multi-cell ROI (GeoMx), or whole transcriptome (Visium HD). Fresh frozen → Discovery path by resolution: subcellular (Stereo-seq, Seq-Scope ★), single-cell (Visium HD, Slide-seqV2), multi-cell (Visium); Hypothesis-driven path → imaging platforms or GeoMx; Protein+RNA → CosMx, Xenium, GeoMx; Rare cells → GeoMx DSP. Quick reference table summarizes recommendations by experimental need.
Landmark Papers Organized by Methodological Approach
1. Sequencing-Based Spatial Arrays (Capturing RNA onto surfaces)
Why these are grouped: These methods work by placing a tissue section onto a slide printed with barcoded capture spots (like beads or DNA nanoballs). The RNA is released from the tissue, captured on the surface, and then sequenced off-slide. The resolution is determined by the physical size and density of the capture spots on the array.
Spatial Transcriptomics Method (The Original) (2016)
Stahl et al. - The foundational paper introducing genome-wide spatial transcriptomics using arrayed barcoded primers on glass slides (the predecessor to 10x Genomics Visium).
- First spatial genome-wide approach
- 100 um diameter spots with positional barcodes
- Compatible with H&E staining for histology overlap
Slide-seq V2 (High-Resolution Beads) (2021)
Stickels et al. - A major improvement in array resolution using tightly packed, tiny microbeads instead of printed spots to achieve near-cellular resolution.
- 10 um bead resolution (near-cellular)
- 10-fold sensitivity improvement over original Slide-seq
- Genome-wide capture
Stereo-seq (Nanoscale Resolution) (2022)
Chen et al. - Utilizing DNA nanoball arrays to achieve the highest resolution among array-based methods, enabling subcellular definitions over large areas.
- Nanoscale resolution (~500 nm center-to-center)
- Very large field of view capability (whole embryos)
- Subcellular resolution with whole transcriptome
2. Imaging-Based In Situ Technologies (Visualizing RNA directly in tissue)
Why these are grouped: Instead of capturing RNA onto a slide, these methods leave the RNA inside the tissue cells. They use fluorescent probes and sophisticated microscopes to capture images of individual RNA molecules over multiple rounds of hybridization. They offer very high (subcellular) resolution but usually target a predefined panel of genes rather than the whole transcriptome.
STARmap (3D In Situ Sequencing) (2018)
Wang et al. - An innovative approach combining hydrogel tissue clearing with error-correcting sequencing directly inside intact tissue blocks.
- Truly 3D intact-tissue RNA sequencing (150 um thick)
- SEDAL sequencing with error correction
- Bypasses reverse transcription using SNAIL probes
seqFISH+ (Transcriptome-Scale Imaging) (2019)
Eng et al. - A landmark paper demonstrating that fluorescence imaging could scale to detect 10,000+ genes at super-resolution using clever color-coding strategies.
- Massive multiplexing (10,000 genes) via pseudocolour barcoding
- Subcellular mRNA localization at super-resolution
- High sensitivity (~35,000 transcripts detected per cell)
CosMx SMI (Commercial High-Plex Imaging) (2022)
He et al. - A representative paper for the new generation of robust, commercial single-molecule imaging platforms suitable for difficult clinical samples.
- High-plex single-molecule imaging (1000+ gene panels)
- Robust performance on FFPE tissue sections
- Focus on clinical translation and reproducibility
3. Beyond 2D RNA: Multi-Modal, 3D, and Temporal Expansions
Why these are grouped: These papers represent the "next frontiers" of spatial biology. They push beyond standard 2D transcriptomics by adding new modalities (like proteins or active translation), analyzing thick 3D tissues, or adding the fourth dimension of time (live-cell tracking).
SPOTS (Spatial Protein & Transcriptome) (2023)
Ben-Chetrit et al. - A pioneering method combining whole transcriptome spatial profiling (Visium) with protein detection in the exact same tissue section.
- True multi-modal integration (RNA + Protein)
- Uses DNA-barcoded antibodies readable by sequencing
- Allows correlation of gene expression vs. actual protein levels
Deep-STARmap & Deep-RIBOmap (3D & Translatomics) (2025)
Sui et al. - Advances 3D imaging into thicker tissues more cheaply, and introduces "translatomics"—mapping actively translating ribosomes, not just total mRNA.
- Scalable 3D profiling in thick blocks (up to 200 um)
- Significant cost reduction via CNVK photocrosslinking
- First spatial translatomics method (RIBOmap)
ESPRESSO (Spatiotemporal Live-Cell Omics) (2025)
Scipioni et al. - Moves from static snapshots to dynamic movies. It uses organelle phenotyping to track cell states over 24+ hours in live cells.
- Adds the temporal dimension (4D tracking)
- Hyperspectral imaging and CNN enhancement
- Tracks live-cell state transitions in 2D and 3D spheroids
4. Benchmarking, Biological Application, and Future Outlook
Why these are grouped: These papers are less about inventing a new method and more about making sense of the existing ones. They include direct head-to-head comparisons of platforms, deep biological applications demonstrating real-world utility, and high-level perspectives on where the field is headed.
Visium-GeoMx-Chromium Comparison (Benchmarking) (2025)
Dong et al. - A crucial resource for users trying to choose a platform. It provides a comprehensive comparison of three major commercial technologies on difficult FFPE tumor samples.
- Compares unbiased sampling (Visium) vs. targeted enrichment (GeoMx)
- Validates FFPE compatibility across major platforms
- Highlights the importance of computational deconvolution
Spatial TRM Cell Diversity (Biological Application) (2025)
Reina-Campos et al. - A prime example of how these tools are used for biological discovery. It integrates multiple spatial platforms to reveal how tissue architecture dictates immune cell behavior.
- Advanced multi-platform integration (Xenium, MERSCOPE, VisiumHD)
- Development of a new tissue coordinate system (IMAP)
- Links spatial niches to immune cell maintenance
Human Cell Atlas Perspective (Future Outlook) (2024)
Rood et al. - A high-level view of how spatial data will be integrated into global efforts like the Human Cell Atlas to create AI-driven foundation models of biology.
- Integrating spatial maps with single-cell census data
- The role of spatial data in understanding development and disease
- The future of AI/ML approaches for atlas-scale analysis
Common Analysis Workflows
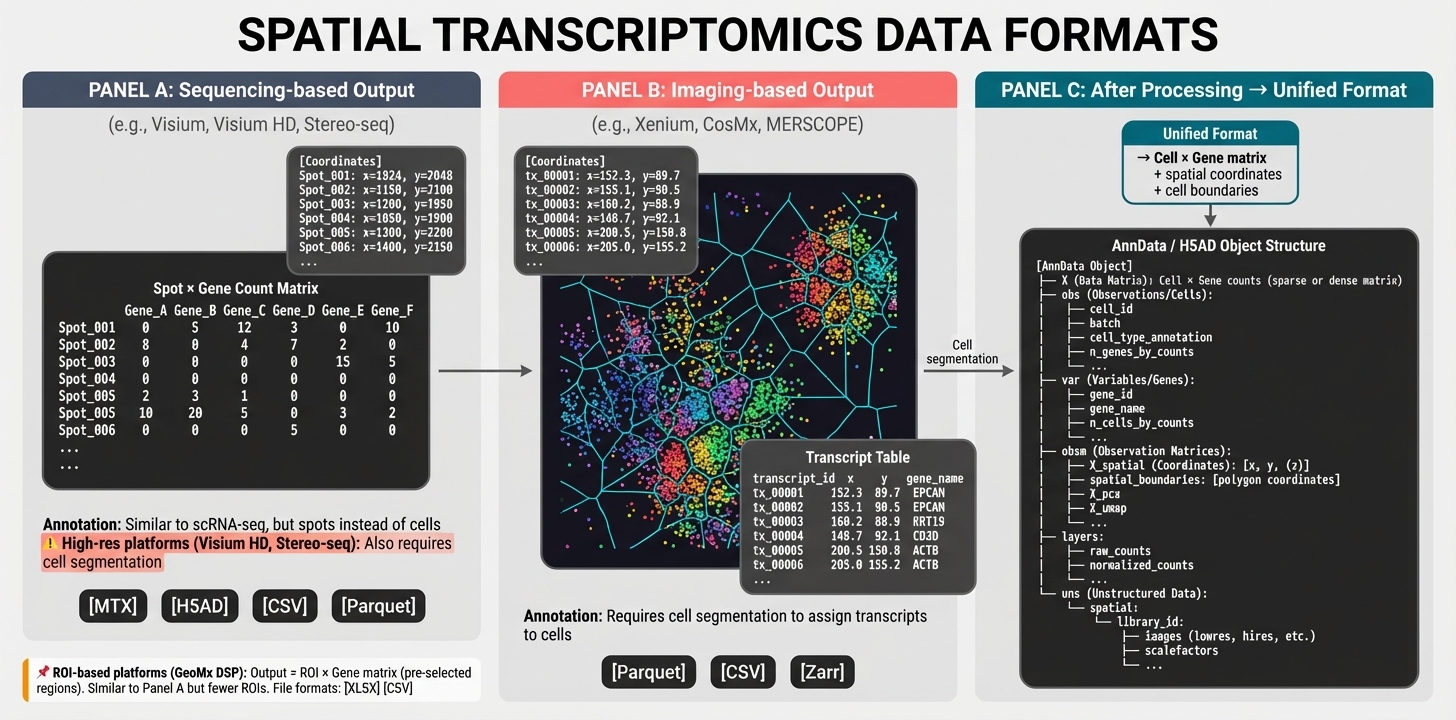
Figure 6. Data output formats across platforms. Panel A (Sequencing-based): Spot × Gene count matrix with coordinates; high-res platforms require segmentation (⚠). Formats: MTX, H5AD, Parquet. Panel B (Imaging-based): transcript coordinate table (transcript_id, x, y, gene); requires cell segmentation. Formats: Parquet, CSV, Zarr. Panel C (Unified): all platforms converge to Cell × Gene matrix with spatial coordinates in AnnData/H5AD structure containing X (counts), obs (cell metadata), var (gene metadata), obsm (spatial coordinates), layers, and uns (images).
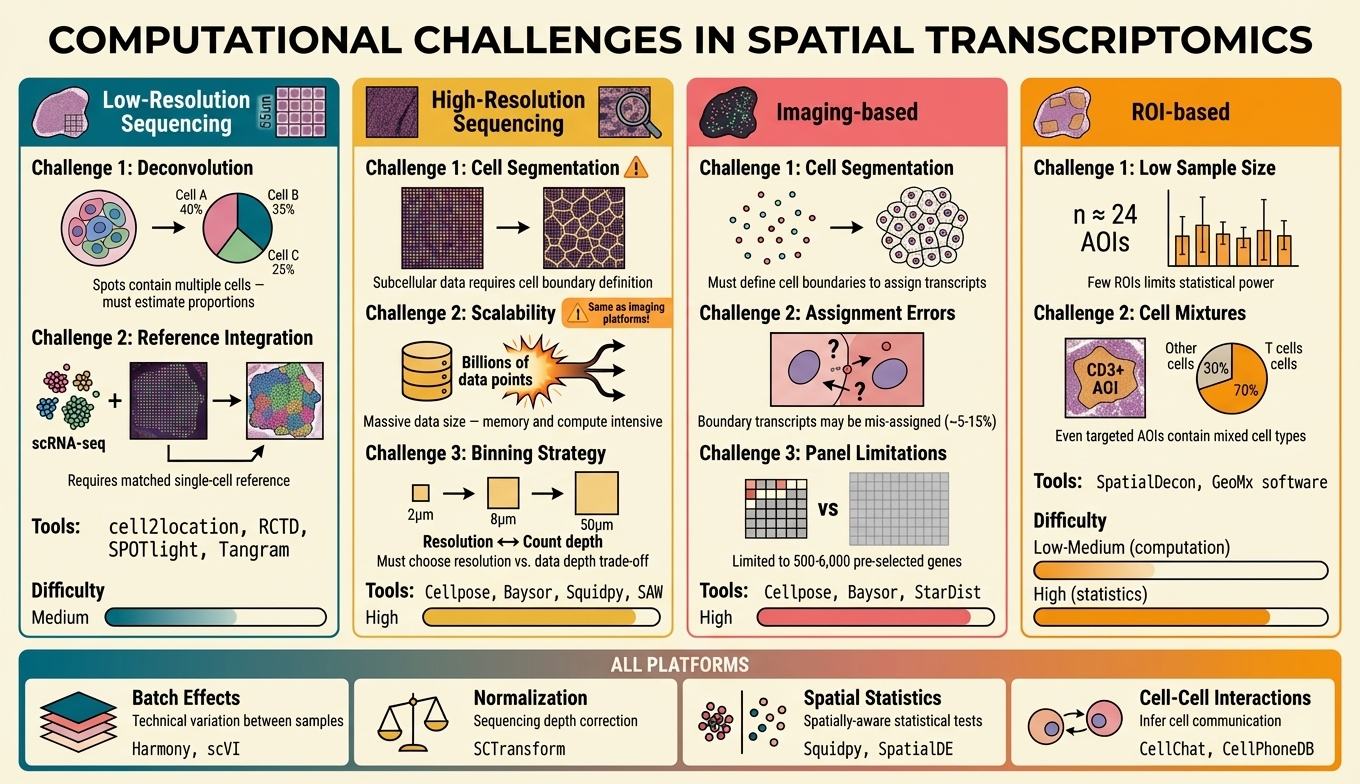
Figure 7. Platform-specific computational challenges. Low-res Sequencing (Visium): deconvolution, reference integration; tools: cell2location, RCTD; difficulty: medium. High-res Sequencing (Visium HD, Stereo-seq): cell segmentation (⚠), scalability, binning strategy; tools: Cellpose, Baysor; difficulty: high. Imaging-based: cell segmentation, assignment errors (~5-15%), panel limitations; tools: Cellpose, Baysor, StarDist; difficulty: high. ROI-based (GeoMx): low sample size, cell mixtures in AOIs; difficulty: low-medium (computation), high (statistics). Shared challenges (all platforms): batch effects, normalization, spatial statistics, cell-cell interactions.
1. Data Processing & Quality Control
- Image Registration: Align multi-round images using fiducial markers or amplicon positions
- Deconvolution: Enhance resolution using PSF-based methods (Huygens, deconvolution algorithms)
- Spot/Barcode Calling: Detect transcripts using local maxima detection, barcode decoding with error correction
- Cell Segmentation: Watershed, CellPose, or Baysor for single-cell boundaries
2. Cell Type Annotation
- Reference-Based: Transfer labels from scRNA-seq using FuseMap, Harmony, or Cell2location
- Unsupervised: Louvain/Leiden clustering on spatial features followed by marker gene validation
- Deconvolution: SpatialDecon, RCTD, or Cell2location for spot-level cell type mixtures
3. Spatial Analysis
- Neighborhood Analysis: Squidpy for cellular interaction scores, Delaunay triangulation for neighbors
- Spatial Domains: BayesSpace, HMRF, or SpaGCN for identifying tissue regions
- Ligand-Receptor: CellChat, CellPhoneDB for cell-cell communication inference
- 3D Analysis: Distance transform watershed, volumetric clustering, spatial statistics
Key Software Packages
| Package | Language | Primary Use |
|---|---|---|
| Scanpy/Squidpy | Python | Single-cell and spatial analysis |
| Seurat | R | Comprehensive spatial workflows |
| Cell2location | Python | Cell type deconvolution |
| CellPose | Python | Cell segmentation |
| Baysor | Julia | Transcript-based segmentation |
| FuseMap | Python | Spatial atlas integration |
| BayesSpace | R | Spatial clustering enhancement |